In my previous work, I focused on many projects, including metagenomics assembly, drug design, multi-omics, AI model building, and software and database development.
-
SApredict. We build the Machine learning model to predict the SA based on the metagenomics, under review.
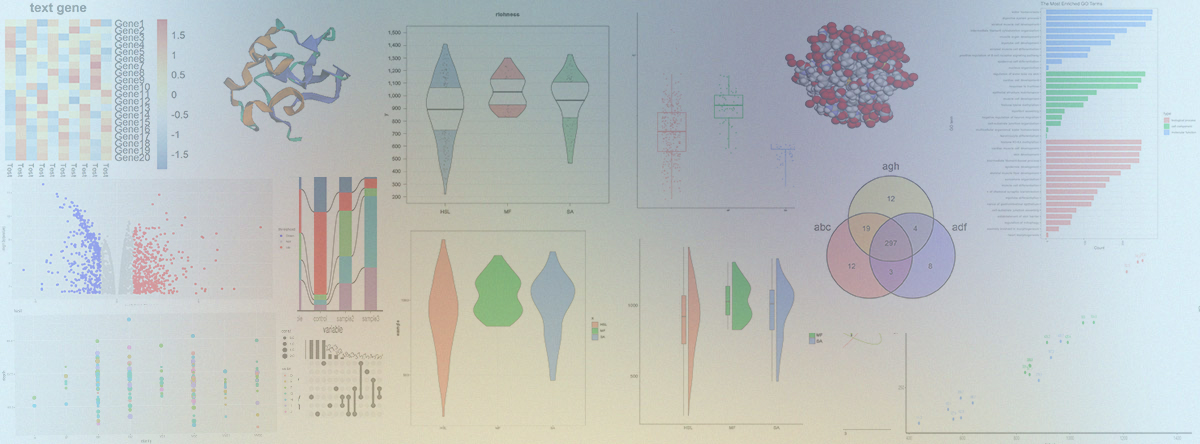
- MicrobioSee. The development of a Multi-omics visualization toolkit.
- Avalable at: https://microbiosee.gxu.edu.cn/
- In this work, we developed MicrobioSee, a real-time interactive visualization tool based on web technologies, which could visualize microbial multi-omics data.
- It includes 17 modules surrounding the major omics data of microorganisms, such as the transcriptome, metagenome, and proteome.
- Most of the tools in MicrobioSee were built with shiny services to achieve real-time interactive plotting.
Cite:
Jinhui Li, Yimeng Sang, Sen Zeng, Shuming Mo, Zufan Zhang, Sheng He, Xinying Li, Guijiao Su, Jianping Liao, and Chengjian Jiang. MicrobioSee: a web-based visualization toolkit for multi-omics of microbiology. Frontiers in Genetics. 2022. 13: 853612. DOI: 10.3389/fgene.2022.853612.
- SMDB. Sulfur metabolism in subtropical marine mangrove sediments fundamentally differs from other habitats, as revealed by SMDB.
- Available at: https://smdb.gxu.edu.cn/
- We introduce SMDB, a manually curated database of sulfur genes based on an in-depth review of the scientific literature and orthology database.
- The SMDB contained a total of 175 genes and covered 11 sulfur metabolism processes with 395,737 representative sequences affiliated with 110 phyla and 2340 genera of bacteria/archaea.
- SMDB will efficiently assist researchers in efficiently analyzing genes of the sulfur cycle from the metagenomic.
Cite:
Shuming Mo, Bing Yan, Tingwei Gao, Jinhui Li, Muhammad Kashif, Jingjing Song, Lirong Bai, Dahui Yu, Jianping Liao & Chengjian Jiang. Sulfur metabolism in subtropical marine mangrove sediments fundamentally differs from other habitats, as revealed by SMDB. Scientific Reports. 2023.13(1): 8126. doi: https://doi.org/10.1038/s41598-023-34995-y

Fulture
Computational biology research is very exciting.
- Now, I focus on exploring the relationship between the repeat areas in multiple genomics and diseases, such as AD.
- More and more species genomics has been sequenced, but few are explored clearly. Using new methods to explore pathogenicity of the repeats.
- At the same time, I want to develop some tools to detect the repeats in multiple species genomics. Machine learning and Deep learning are very useful algorithms for that. GPU CUDA technique can also promote my research.
- Also, I want to continue my metagenome research.
- Developing software to assemble the metagenomics with low memory is necessary now.
- Based on the FMT technique(Fecal microbiota transplantation), it is very meaningful to improve the well-being of cancer or sub-healthy people.